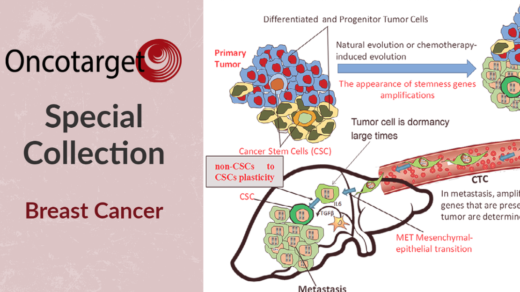
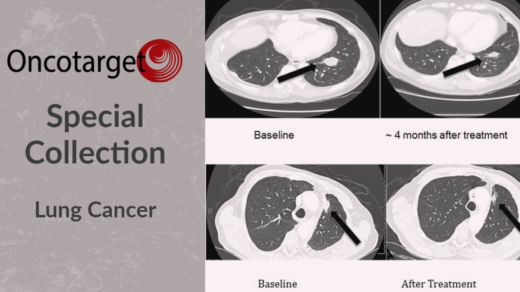
Researchers investigated common gut microbiome features of response among immunotherapy patients with different advanced-stage cancers.
Immunotherapy has become a powerful breakthrough in cancer treatment, however, 50% of patients do not respond to immunotherapy. What internal or external features inhibit or confer patient or tumor response to immune system-harnessing therapeutics? These patient/tumor features that impact responsiveness to immunotherapy have yet to be fully elucidated.
“Increasing evidence has emerged that gut microbial communities help shape the host immune system [9–11].”
The Gut Microbiome
The gut microbiome is the collection of all microbes living in the gastrointestinal tract. A growing body of evidence indicates that the gut microbiome is an important component of the human immune system. This evidence has prompted researchers to hypothesize that the gut microbiome may play a role in immunotherapy response. It has since become a focal point in immunotherapy research and also has the potential to serve as a biomarker for immunotherapy response.
Researchers previously found evidence that specific gut bacteria can influence immunotherapy outcomes by modulating immune responses in patients with melanoma, non–small cell lung cancer and renal cell cancer. (“Treatment responders generally exhibit increased gut microbial community diversity and are enriched in certain bacterial taxa including Akkermansia and Bifidobacterium [16, 19].”) While this is important, it is unclear whether these response signals are generalizable across different tumor types, geographical variations in the microbiome or sequencing platforms and analysis methodologies. Identifying microbiome features associated with immunotherapy response regardless of the type of cancer or where it started in the body may be the next step toward improving immunotherapy outcomes in cancer patients.
The Study
In a new study published in Oncotarget, Hai Liang, Jay-Hyun Jo, Zhiwei Zhang, Margaret A. MacGibeny, Jungmin Han, Diana M. Proctor, Monica E. Taylor, You Che, Paul Juneau, Andrea B. Apolo, John A. McCulloch, Diwakar Davar, Hassane M. Zarour, Amiran K. Dzutsev, Isaac Brownell, Giorgio Trinchieri, James L. Gulley, and Heidi H. Kong, from the National Institutes of Health Library, National Cancer Institute (NCI), National Human Genome Research Institute, West Virginia University, Zimmerman Associates Inc., and the University of Pittsburgh, used machine learning models to analyze gut microbiomes and predict patient response to immunotherapy. On July 19, 2022, the researchers published their paper in Oncotarget’s Volume 13, entitled, “Predicting cancer immunotherapy response from gut microbiomes using machine learning models.”
To find common gut microbiome features of response to immunotherapy, a tumor-agnostic and geographically limited (U.S. patients only) approach was used in this study. The researchers’ discovery cohort included 16 patients with nine different advanced-stage cancers who were enrolled in NCI immunotherapy trials (the NCI cohort). Patient samples and 16S rRNA gene sequencing data were collected. Patients who positively responded to immunotherapies (responders) and those who did not (non-responders) were categorized. The researchers used uni- and multivariate analyses to identify common microbiome features and complex microbial community interactions.
Common microbiome features and immunotherapy response signals in the discovery cohort were further validated with larger datasets. Three previously published 16S rRNA gene sequencing datasets from melanoma patients were added to a combined dataset. This combined dataset was used to validate the NCI cohort results in a meta-analysis. Data from all four studies were used in statistical analyses and machine learning models aimed to predict immunotherapy response.
“Using the combined dataset, we trained and validated models with machine learning algorithms to predict patients’ clinical responses, followed by cross-sequencing-platform validation using shotgun metagenomic sequencing data.”
The Results
“Results suggest baseline gut microbiome features may be predictive of clinical outcomes in oncology patients on immunotherapies, and some of these features may be generalizable across different tumor types, patient cohorts, and sequencing platforms.”
The researchers found that the gut microbiome composition of responders to immunotherapy was different from that of non-responders. Several species of bacteria were differentially abundant between responders and non-responders in the NCI cohort, and some were also consistent with results from the meta-analysis. Hierarchical clustering showed a higher immunotherapy response rate among patients with enriched bacteria in the Firmicutes phylum and a lower response rate among patients enriched in the Bacteroidetes phylum at baseline. Machine learning models using microbiome features and immunotherapy response signals found in this study demonstrated a favorable prediction accuracy with the highest AUC (area under the curve) value of around 0.75.
Conclusion
“In conclusion, analyses of our cohort and the combined microbiome dataset have provided a robust assessment of immunotherapy patients’ gut microbiomes.”
Researchers used a tumor-agnostic approach to find common gut microbiome features of response to immunotherapy in cancer patients. The machine learning models developed in this study were able to demonstrate a favorable prediction accuracy of 75% at best. These results suggest that the gut microbiome may be predictive of immunotherapy response in cancer patients and that some of these features may be generalizable across different tumor types, patient cohorts, and sequencing platforms. These findings suggest that the gut microbiome is a promising biomarker for immunotherapy response. However, more research is needed to validate these findings in larger and more diverse patient populations.
“Results suggest baseline gut microbiome features may be predictive of clinical outcomes in oncology patients on immunotherapies, and some of these features may be generalizable across different tumor types, patient cohorts, and sequencing platforms. Findings demonstrate how machine learning models can reveal microbiome-immunotherapy interactions that may ultimately improve cancer patient outcomes.”
Click here to read the full research paper published by Oncotarget.
ONCOTARGET VIDEOS: YouTube | LabTube | Oncotarget.com
—
Oncotarget is an open-access journal that publishes primarily oncology-focused research papers in a continuous publishing format. These papers are available at no cost to readers on Oncotarget.com. Open-access journals have the power to benefit humanity from the inside out by rapidly disseminating information that may be freely shared with researchers, colleagues, family, and friends around the world.
For media inquiries, please contact media@impactjournals.com.