In a new Oncotarget editorial, researchers discussed their study on CD56 in multiple myeloma.
The Trending With Impact series highlights Oncotarget publications attracting higher visibility among readers around the world online, in the news, and on social media—beyond normal readership levels. Look for future science news about the latest trending publications here, and at Oncotarget.com.
—
Multiple myeloma (MM) is a type of blood cancer that affects plasma cells in the bone marrow. These plasma cells, which are responsible for producing antibodies, become abnormal and begin to grow uncontrollably. This results in a buildup of abnormal cells in the bone marrow, leading to decreased production of healthy blood cells, bone damage and a host of other symptoms. MM is incredibly heterogenic, and this variability often leads to unsatisfactory long-term treatment outcomes in many patients with MM. Targets for new treatments and biomarkers of response are needed to improve patient outcomes.
In a new editorial paper published in Oncotarget, researchers Francesca Cottini and Don Benson from The Ohio State University discuss a 2022 research paper they co-authored in which CD56 (or neuronal cell adhesion molecule; NCAM1) was thoroughly described as a biomarker and therapeutic target in multiple myeloma. On January 26, 2023, their editorial about this research paper was published in Oncotarget, entitled, “To be or not to be: the role of CD56 in multiple myeloma.”
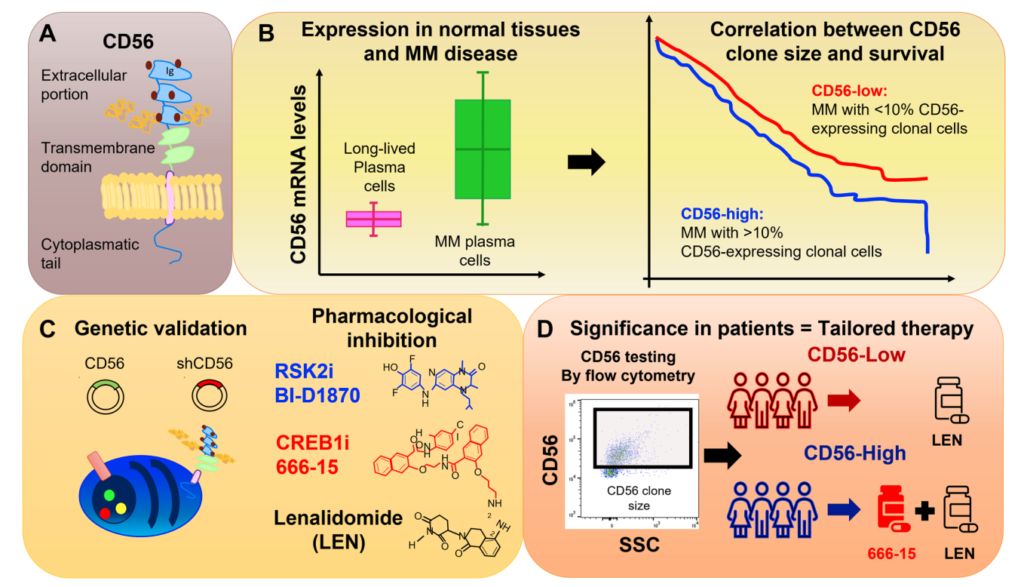
“Among others, CD56 is present at variable levels in approximately 70% of patients with multiple myeloma; however, very little is known about CD56 role in multiple myeloma.” (2022 Cottini et al.)
CD56 in MM
The role of CD56 in multiple myeloma is a topic of ongoing research and discussion among scientists and medical professionals. CD56 is a protein that is found on the surface of many different cell types, including plasma cells. Researchers have demonstrated that it plays a key role in the development and progression of multiple myeloma, making it a potential target for new treatments.
One of the main functions of CD56 is to regulate the growth and survival of plasma cells. In normal cells, CD56 helps to prevent uncontrolled growth and division of cells. However, in multiple myeloma cells, CD56 appears to play a different role. Research has shown that CD56 is overexpressed in multiple myeloma cells, leading to an increase in cell growth and division.
Additionally, CD56 has been found to play a role in the immune system’s response to cancer cells. In multiple myeloma, CD56 can suppress the immune system’s response to the abnormal cells, allowing them to continue growing unchecked. This is why multiple myeloma is often resistant to traditional cancer treatments such as chemotherapy and radiation.
Targeting CD56 in MM
There are currently several strategies being explored to target CD56 in multiple myeloma. One approach is to use drugs that block the function of CD56, in order to prevent it from promoting cell growth and division. Another approach is to use immunotherapies that stimulate the immune system to attack the abnormal cells. This can help to overcome the suppression of the immune system by CD56 and lead to more effective cancer treatment.
There is also research being conducted into the use of CAR T-cell therapy. CAR T-cell therapy involves genetically modifying a patient’s own immune cells to attack the cancer cells. In this type of therapy, the immune cells are modified to target CD56 specifically, which allows them to attack and destroy the multiple myeloma cells more effectively.
Cottini et al.
In this editorial, the researchers discuss their study, which looked at CD56-expressing clonal MM cells in more than 700 patients at the time of MM diagnosis. The researchers found that the size of these cells varied between patients and increased as the disease worsened. Results demonstrated that having a large amount of these cells was linked to worse outcomes and shorter responses to treatment.
The study then looked at how changing the expression of CD56 affected the behavior of MM cells and found that it influenced cell growth and survival. They also discovered that a protein called RSK2 and another called CREB1 play a role in this process. They then tested medicines to block these proteins and found that they were effective in killing MM cells that had a high amount of CD56, but not as much in those with low levels of CD56.
“The authors’ preclinical data support the use of synthetic lethal approaches by CREB1/RSK2 inhibition in combination with lenalidomide, as a strategy to overcome CRBN downregulation in CD56-high MM.”
Conclusion
“In summary, this study provides a detailed description of CD56 role in MM, opening new clinically relevant scenarios.”
The role of CD56 in multiple myeloma is complex and still not fully understood. However, the researchers who wrote this editorial aimed to clearly define CD56’s key role in the development and progression of this disease, making it a potential target for new treatments. By better understanding the function of CD56, scientists and medical professionals can continue to develop new and innovative therapies to improve the lives of those affected by multiple myeloma.
“Since the majority of clinical laboratories have the capability to perform CD56 staining and define a threshold of positivity, CD56 expression can be both a prognostic and predictive factor of response to therapies, an unmet need in the MM field (Figure 1).”
Click here to read the full editorial published in Oncotarget.
ONCOTARGET VIDEOS: YouTube | LabTube | Oncotarget.com
—
Oncotarget is an open-access, peer-reviewed journal that has published primarily oncology-focused research papers since 2010. These papers are available to readers (at no cost and free of subscription barriers) in a continuous publishing format at Oncotarget.com. Oncotarget is indexed/archived on MEDLINE / PMC / PubMed.
For media inquiries, please contact media@impactjournals.com.