“Precision medicine is an innovative approach to disease prevention and treatment that considers differences in people’s genes, injuries, environments, and lifestyles to target the right therapies to the right patients at the right time.”
Could a deeper understanding of one of the deadliest lung cancers lead to more effective treatments? Recent research offers a promising way forward, aiming to improve patient outcomes and provide clinicians with valuable insights.
Small Cell Lung Cancer (SCLC) is a particularly aggressive form of lung cancer. It spreads fast and does not always respond well to conventional therapies such as chemotherapy. Although SCLC accounts for around 15% of all lung cancer cases, survival rates are extremely low. Only less than 5% of patients live more than five years after diagnosis. These alarming statistics highlight the critical need for new treatments. A team of researchers from the Federal University of Ceará, working together with collaborators from Argentina and Spain, may have found part of the solution.
The Study: Finding Clues in Tumor Biomarkers
Published in Oncotarget Volume 15 on October 11, 2024, the study is titled “Relationship between the expressions of DLL3, ASC1, TTF-1 and Ki-67: First steps of precision medicine at SCLC.” Led by corresponding author, Dr. Fabio Tavora, the research team analyzed tumor samples from 64 adult patients diagnosed with SCLC between 2022 and 2024. Their focus was on specific biomarkers, molecules that reveal the unique biology of a tumor, and that could lead to better ways to improve diagnosis and treatment for SCLC.
The Challenge: Why Current Treatments Are Not Enough
Current treatments for SCLC, like chemotherapy, are based on a “one size fits all” approach. While chemotherapy can initially decrease tumor size, its effects are often temporary, in addition to the harsh side effects that the patients can experience. This type of universal treatment overlooks each tumor’s specific biological characteristics, which is one of the main reasons chemotherapy has such low long-term success rates for SCLC patients.
To address this problem, the researchers studied the presence of distinct biomarkers. Since these molecules usually differ from tumor to tumor, they can provide valuable insights into tumor behavior. The goal was to discover new ways to personalize treatments, making them more efficient.
Technology: Using Digital Tools for Precision
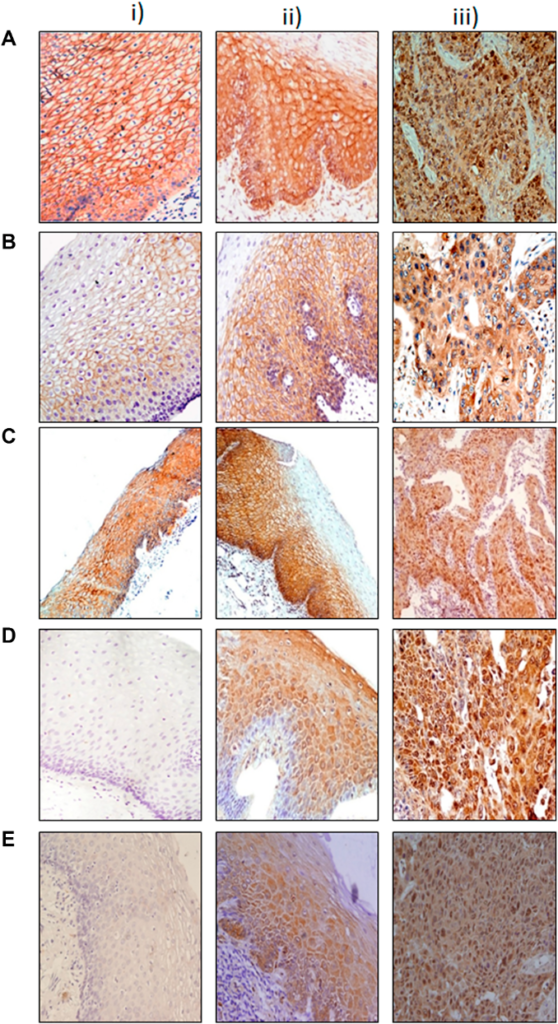
In addition to standard techniques like immunohistochemistry, the researchers used QuPath, a novel digital pathology tool. QuPath enabled the researchers to evaluate tumor samples with unprecedented precision and resolution, revealing differences in biomarker expression between patients and between tumors, thus demonstrating that no two tumors are identical. This highlighted the importance of tailored treatment options.
The Results: Two Important Biomarkers
The study revealed two key biomarkers with the potential to improve SCLC treatment: Delta-like ligand 3 (DLL3) and Thyroid Transcription Factor-1 (TTF-1).
The Breakthrough: DLL3 and TTF-1 as Game-Changing
DLL3, a protein identified almost exclusively on the surface of SCLC tumor cells, has emerged as a potential therapeutic target. In this study, DLL3 was found in more than 70% of the SCLC tumors and its expression was tumor-specific. This finding makes it a suitable target for precision medicine, as treatments can target DLL3-positive cells while avoiding healthy tissue.
TTF-1, traditionally used as a diagnostic marker for lung cancer, showed potential as a prognostic biomarker in this study. Patients with TTF-1-positive tumors demonstrated better survival rates. The study also discovered a link between the expressions of TTF-1 and DLL3, suggesting that these two biomarkers could be used together to help choose treatments.
Therapeutic Potential: Toward Personalized Treatments
Unlike traditional therapies, which take a generic approach, biomarker-based treatments are tailored to the unique characteristics of each tumor. This personalization could make treatments more effective while reducing the side effects that patients experience.
Samuel Silva, the study’s first author, during an interview, emphasized the importance of the relationship between TTF-1 and DLL3. TTF-1 serves as both diagnostic and survival prediction tool, while DLL3 creates new opportunities for targeted therapy. Together, these biomarkers offer hope for more effective and individualized treatments for SCLC.
One promising development is the recent FDA approval of Tarlatamab, a therapeutic compound that uses the body’s immune system to target and destroy DLL3-positive tumor cells. This is an example of how biomarkers like DLL3 can be successfully translated into clinical settings.
Looking Ahead: What’s Next for SCLC Research
While this study represents a significant step forward on SCLC treatment, there is still more work to be done. In his interview, Silva explained that his team plans “… to look into the genomics and transcriptomics landscapes and the interactions of all these molecules,” aiming to better understand the molecular mechanisms driving SCLC and to further refine the therapeutic strategies.
Future clinical trials will be necessary to validate these findings and assess how effective and reliable DLL3 and TTF-1 are in guiding treatment for SCLC patients. With continued research, the ultimate goal is to make precision medicine a standard option for all SCLC patients.
Conclusion
SCLC is one of the most challenging cancers to treat, but this study offers hope. QuPath played a key role in showing how biomarkers like DLL3 and TTF-1 can guide more personalized and effective treatments. By focusing on these biomarkers, researchers are leading the way for better ways to diagnose and treat this aggressive disease. Precision medicine, which tailors treatments to each patient’s unique needs, promises to improve survival rates and quality of life for those facing SCLC.
Click here to read the full research paper in Oncotarget.
—
Oncotarget is an open-access, peer-reviewed journal that has published primarily oncology-focused research papers since 2010. These papers are available to readers (at no cost and free of subscription barriers) in a continuous publishing format at Oncotarget.com.
Oncotarget is indexed and archived by PubMed/Medline, PubMed Central, Scopus, EMBASE, META (Chan Zuckerberg Initiative) (2018-2022), and Dimensions (Digital Science).
Click here to subscribe to Oncotarget publication updates.
For media inquiries, please contact media@impactjournals.com.