Researchers studied the dynamic behavior of gene expression during the development of endocrine therapy resistance in breast cancer.
The Trending With Impact series highlights Oncotarget publications attracting higher visibility among readers around the world online, in the news, and on social media—beyond normal readership levels. Look for future science news about the latest trending publications here, and at Oncotarget.com.
—
Hormones can cause tumor growth in some subtypes of breast cancer. Endocrine therapy, also known as hormone therapy, is a type of cancer treatment that removes or blocks the hormones which fuel breast cancer growth. This treatment is often given as adjuvant therapy after breast cancer surgery to lower the risk of cancer reoccurrence. In some cases, endocrine therapy may be used as a first-line treatment for hormone receptor-positive breast cancers, such as estrogen receptor-positive (ER-positive) breast cancer. However, ER-positive tumors frequently become unresponsive to endocrine therapy, and tumor regrowth can occur after treatment. The underlying causes of endocrine resistance are mostly undetermined.
“Endocrine therapies have been successful at improving cancer outcomes; however, the development of endocrine resistance, or resistance to inhibition of ER actions, remains a roadblock in breast cancer treatment.”
Recently, researchers—from UTHealth Houston, University of Chicago, University of Texas MD Anderson Cancer Center, and the University of Houston—used a new statistical and computational pipeline method of analysis to study the dynamic behavior of gene expression during the development of endocrine resistance in breast cancer. Their trending research paper published in Oncotarget on April 06, 2022, is entitled, “A novel group of genes that cause endocrine resistance in breast cancer identified by dynamic gene expression analysis.”
The Pipeline
“In this study, we explored the dynamic behavior of the entire gene population to identify novel genes that play fundamental roles in the development and progression of endocrine-resistant breast cancer.”
Pipeline analysis in biology is a method of studying and analyzing a group of genes or proteins in order to understand their structure and function. The pipeline can be used to determine gene dynamics, clusters, similarities, and networks. In this case, the researchers used it to understand how endocrine resistance develops over time.
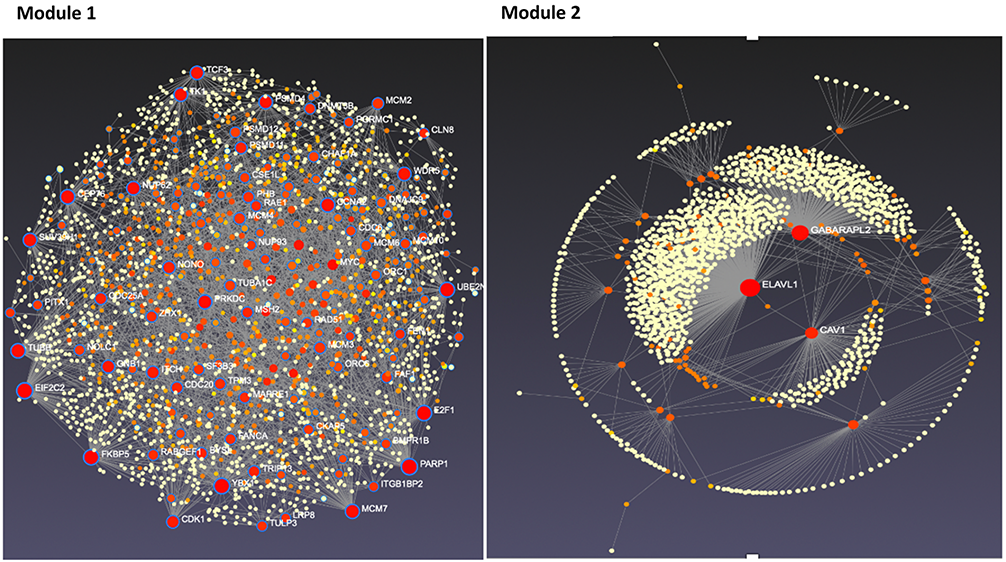
“The pipeline provides three main functions. First, statistical hypothesis testing determines a set of dynamic response genes (DRGs) that exhibit significant changes over time. Next, these DRGs are clustered into gene response modules (GRMs), sets of DRGs with similar time course expression patterns. Finally, the GRMs associations and regulatory effect are analyzed as a gene regulatory network using ordinary differential equations.”
The Study
To begin this study, the researchers first aimed to select a cell-based model that represents endocrine resistance in patients as closely as possible. They gathered data from breast cancer patients who were either resistant or sensitive to endocrine therapies and compared them with publicly available gene expression data. Results showed that the LTED MCF7 cell model displayed similar endocrine resistance to patient tumor data.
Next, the researchers observed the development of endocrine therapy resistance in the LTED MCF7 cell model, as well as the changes in gene expression over time. This data was collected and used to develop a mathematical model of gene expression dynamics during endocrine therapy resistance development. After statistical and computational pipeline analysis, the team identified a group of 254 genes whose time course expression significantly changed during the development of endocrine therapy resistance. They then aimed to validate their findings and used multiple bioinformatics approaches to narrow down this group of candidate genes.
“To further refine the genes common to endocrine resistance development and progression, we utilized several bioinformatic approaches designated to rank and prioritize the 254 common genes.”
The Results
Candidate genes were narrowed down to a novel group of 34 genes whose time course expression most significantly changed during LTED MCF7 cell modeling of endocrine-resistant breast cancer development. In addition, microarray analysis also showed that a subset of these genes was differentially expressed in triple-negative breast cancer (TNBC). This suggests that there may be shared genetic mechanisms between endocrine-resistant breast cancer and TNBC.
“As these two subtypes of breast cancer are the most fatal breast cancers with no known effective therapeutic approaches available to date, research on underlying genetic factors is of great importance.”
Conclusion
Their analysis led to the identification of a novel group of 34 genes that may play a role in endocrine resistance. Interestingly, some of these genes were also differentially expressed in TNBC. These findings could potentially lead to the development of new therapeutic strategies to overcome endocrine therapy resistance in some of the most difficult to treat and fatal breast cancers.
“Our analysis identified novel candidate genes with potential significance in endocrine-resistant breast cancer as well as TNBC, which opens new doors for designing novel therapeutic approaches for endocrine-resistant breast cancer and TNBC.”
Click here to read the full research paper published by Oncotarget.
ONCOTARGET VIDEOS: YouTube | LabTube | Oncotarget.com
—
Oncotarget is an open-access journal that publishes primarily oncology-focused research papers in a continuous publishing format. These papers are available at no cost to readers on Oncotarget.com. Open-access journals have the power to benefit humanity from the inside out by rapidly disseminating information that may be freely shared with researchers, colleagues, family, and friends around the world.
For media inquiries, please contact media@impactjournals.com.